RNA detection with FISH-quant¶
FISH-quant¶
FISH-quant is a Matlab toolbox to localize RNAs in 3D from smFISH images.
Detection of individual RNAs¶
Below only a quick summary of the analysis steps is presented. For more details consult the FQ manual..
- Open FQ
- Set analysis folder to folder containing the image that should be analysed:
Menu Folder>Set root folder) - Open image that should be analysed.
- [Optional] Draw outline of embryo / cells / structure of interest. If you omit t his step, RNAs will be detected in the entire image.
- Open dedicated interface: Button
Define outlines - Draw a new cell: Button
New cell - Save outline: Button
Quick-save - Return to main FQ: Button
Finished
- Open dedicated interface: Button
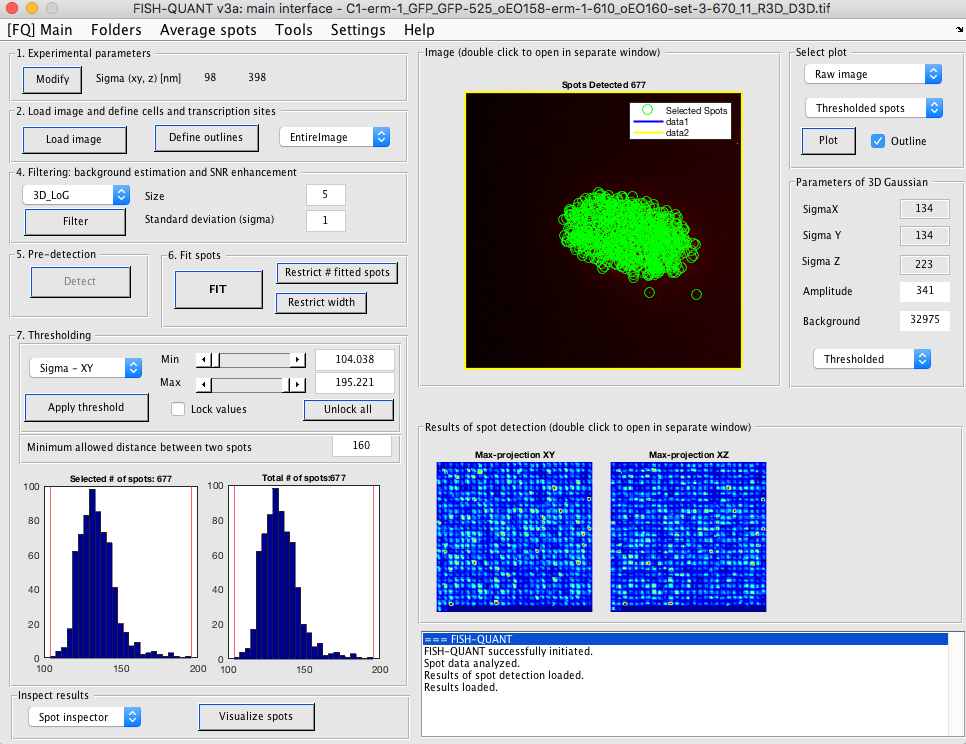
- Filter image (default filter with
LoGworks usually well). - Inspect image. On the right part of the interface select
Filtered image, disableoutline, and double click on the image. This will show a maximum intensity projection of the filtered image in a separate window. Here, you can change the contrast and zoom. In order to determine an appropriate threshold in the next step, determine a generous range of intensities range corresponding to the individual RNA molecules. -
Set pre-detection settings. FQ will test how many RNAs are detected with a range of user-defined intensity thresholds (based on the manual inspection of the image from above).
- Open dedicated interface: Button
Detect - In the user-dialog change the first two parameter: minimum and maximum threshold to test. We usually use a minimum value that’s somewhat lower than the lowest RNAs to also consider intensity value corresponding to background.
- FQ will calculate the number of detected RNAs for a range of values in this interval. Depending on the size of the image, this can take a little while. Once done, a plot with the number of detected RNAs as a function of the different thresholds is shown. If the specified range is not appropriate, it can be changed in this interface the computation be repeated.
- Ideally, this curve shows a plateau for a value range corresponding to a good detection threshold. If that’s not the case, a reasonable value corresponding to a visual assessment of what individual RNAs are should be chosen. A pre-detection with a given detection threshold can be perform and the results be inspected in a separate window.
One other parameter that could be important to adjust is the cropping region around each RNA that is considered for analysis and fitting. Reducing this size allows sometimes to better detect closely spaced RNAs but at the cost of an imprecision in the fit. For most applications a value of +/- 2 voxels in XY and Z are a good compromise.
- Open dedicated interface: Button
-
Once you are satisfied with the settings press on
Perform detection for all cellsto return the main window. - Press button
Fitto fit each detected spot with a 3D Gaussian function. - [Optional]. You can set threshold values for the different fitting parameters, e.g. remove spots with very small or very large standard deviations.
- You can now save the detection settings:
[FQ] Main>Save>Save detection settings. As a file-name specifyFQ__settings_MATURE.txt - If you are satisfied with these detection results, you can save them directly:
[FQ] Main>Save>Detection spots [All].